Outputs
Outputs from Parthenon are controlled via <parthenon/output*> blocks,
where * should be replaced by a unique integer for each block.
To disable an output block without removing it from the input file set
the block’s dt < 0.0.
In addition to time base outputs, two additional options to trigger outputs (applies to HDF5, restart and histogram outputs) exist.
Signaling: If
Parthenoncatches a signal, e.g.,SIGALRMwhich is often sent by schedulers such as Slurm to signal a job of exceeding the job’s allocated walltime,Parthenonwill gracefully terminate and write output files with afinalid rather than a number. This also applies to theParthenoninternal walltime limit, e.g., when executing an application with the-t HH:MM:SSparameter on the command line.File trigger: If a user places a file with the name
output_nowin the working directory of a running application,Parthenonwill write output files with anowid rather than a number. After the output is being written theoutput_nowfile is removed and the simulation continues normally. The user can repeat the process any time by creating a newoutput_nowfile.
Note, in both cases the original numbering of the output will be
unaffected and the final and now files will be overwritten each
time without warning.
HDF5
Parthenon allows users to select which fields are captured in the HDF5
(.phdf) dumps at runtime. In the input file, include a
<parthenon/output*> block, list of variables, and specify
file_type = hdf5. A dt parameter controls the frequency of
outputs for simulations involving evolution. If the optional parameter
single_precision_output is set to true, all variable data will
be written in single precision. A <parthenon/output*> block might
look like
<parthenon/output1>
file_type = hdf5
write_xdmf = true # Determines whether xdmf annotations are output
# nonexistent variables/swarms are ignored
variables = density, velocity, & # comments are still ok
energy # notice the & continuation character
# for multiline lists
swarms = tracers, photons # Particle swarms
swarm_variables = x, y, z # swarm variables output for every swarm
# Each swarm can sepcify in a separate list which additional
# variables it would like to output.
tracers_variables = x, y, z, rho, id
photons_variables = x, y, z, frequency
dt = 1.0
file_number_width = 6 # default: 5
use_final_label = true # default: true
# Sparse variables may not be allocated on every block. By default
# parthenon outputs de-allocated variables as 0 in the output
# file. However, it is often convenient to output them as NaN
# instead, marking deallocated and allocated but zero as
# separate. This flag turns this functionality on.
sparse_seed_nans = false # default false
This will produce an hdf5 (.phdf) output file every 1 units of
simulation time containing the density, velocity, and energy of each
cell. The files will be identified by a 6-digit ID, and the output file
generated upon completion of the simulation will be labeled
*.final.* rather than with the integer ID.
HDF5 and restart files write variable field data with inline compression
by default. This is especially helpful when there are sparse variables
allocated only in a few blocks, because all other blocks would write
zeros of these variables, which can drastically increase output file
size (and decrease I/O performance) without compression. The optional
parameter hdf5_compression_level can be used to set the compression
level (between 1 and 9, default is 5). If parthenon is compiled with
support for compression, this also enables (logical) chunking of the
data in blocks of nx1*nx2*nx3. Compression (and thus chunking) can
be disabled altogether with the CMake build option
PARTHENON_DISABLE_HDF5_COMPRESSION.
See the Building Parthenon for more details.
Tuning HDF5 Performance
Tuning IO parameters can be passed to Parthenon through the use of environment variables. Available environment variables are:
Environment Variable |
Initial State |
Value Type |
Description |
|---|---|---|---|
H5_sieve_buf_size
H5_meta_block_size
H5_alignment_threshold
H5_alignment_alignment
H5_defer_metadata_flush
MPI_access_style
MPI_collective_buffering
MPI_cb_block_size
MPI_cb_buffer_size
|
disabled
disabled
disabled
disabled
disabled
enabled
disabled
N/A
N/A
|
int
int
int
int
int
string
int
int
int
|
Sets the maximum size of the data sieve buffer, in bytes. The value should be equal to a multiple of the disk block size. If no value is set then the default is 256 KiB.
Sets the minimum metadata block size, in bytes. If no value is set then the default is 8 MiB. May help performance if enabled.
The threshold value, in bytes, of H5Pset_alignment. Setting to 0 forces everything to be aligned. If a value is not set then the default is 0. Setting the environment variable automatically enables alignment.
The alignment value, in bytes, of H5Pset_alignment. If a value is not set then the default is 8 MiB. Setting the environment variable automatically enables alignment. H5Pset_alignment sets the alignment properties of a file access property list. Choose an alignment that is a multiple of the disk block size, enabling this usually shows better performance on parallel file systems. However, enabling may increase the file size significantly.
Value of 1 enables deferring metadata flush. Value of 0 disables. Experiment with before using.
Specifies the manner in which the file will be accessed until the file is closed. Default is “write_once”
Value of 1 enables MPI collective buffering. Value of 0 disables. Experiment with before using.
Sets the block size, in bytes, to be used for collective buffering file access. Default is 1 MiB.
Sets the total buffer space, in bytes, that can be used for collective buffering on each target node, usually a multiple of cb_block_size. Default is 4 MiB.
|
Restart Files
Parthenon allows users to output restart files for restarting a simulation. The restart file captures the input file, so no input file is required to be specified. Parameters for the input can be overridden in the usual way from the command line. At a future date we will allow for users the ability to extensively edit the parameters stored within the restart file.
In the input file, include a <parthenon/output*> block and specify
file_type = rst. A dt parameter controls the frequency of
outputs for simulations involving evolution. A <parthenon/output*>
block might look like
<parthenon/output7>
file_type = rst
dt = 1.0
This will produce an hdf5 (.rhdf) output file every 1 units of
simulation time that can be used for restarting the simulation.
To use this restart file, simply specify the restart file with a
-r <restart.rhdf> at the command line. If both -r <restart.rhdf>
and -i <input.in> are specified, the simulation will be restarted from
the restart file with input parameters updated (or added) from the input file.
For physics developers: The fields to be output are automatically
selected as all the variables that have either the Independent or
Restart Metadata flags specified. No other intervention is
required by the developer.
History Files
In the input file, include a <parthenon/output*> block and specify
file_type = hst. A dt parameter controls the frequency of
outputs for simulations involving evolution. The default behavior is to provide
all enrolled history outputs, but output can be limited to a specific set of
packages with an optional comma-separated list argument
packages = package_a, package_b. A <parthenon/output*>
block might look like
<parthenon/output8>
file_type = hst
dt = 1.0
packages = advection_app
This will produce a text file (.hst) output file every 1 units of
simulation time. The content of the file is determined by the functions
enrolled by specific packages, see History output.
Histograms
Parthenon supports calculating flexible 1D and 2D histograms in-situ that are written to disk in HDF5 format. Currently supported are
1D and 2D histograms (see examples below)
binning by variable or coordinate (x1, x2, x3 and radial distance)
counting samples and or summing a variable
weighting by volume and/or variable
The output format follows numpy convention, so that plotting data
with Python based machinery should be straightforward (see example below).
In other words, 2D histograms use C-ordering corresponding to [x,y]
indexing with y being the fast index.
In general, histograms are calculated using inclusive left bin edges and
data equal to the rightmost edge is also included in the last bin.
A <parthenon/output*> block containing one simple and one complex
example might look like:
<parthenon/output8>
file_type = histogram # required, sets the output type
dt = 1.0 # required, sets the output interval
hist_names = myname, other_name # required, specifies the names of the histograms
# in this block (used a prefix below and in the output)
# 1D histogram ("standard", i.e., counting occurance in bin)
myname_ndim = 1
myname_x_variable = advected
myname_x_variable_component = 0
myname_x_edges_type = log
myname_x_edges_num_bins = 10
myname_x_edges_min = 1e-9
myname_x_edges_max = 1e0
myname_binned_variable = HIST_ONES
# 2D histogram of volume weighted variable according to two coordinates
other_name_ndim = 2
other_name_x_variable = HIST_COORD_X1
other_name_x_edges_type = list
other_name_x_edges_list = -0.5, -0.25, 0.0, 0.25, 0.5
other_name_y_variable = HIST_COORD_X2
other_name_y_edges_type = list
other_name_y_edges_list = -0.5, -0.1, 0.0, 0.1, 0.5
other_name_binned_variable = advected
other_name_binned_variable_component = 0
other_name_weight_by_volume = true
other_name_weight_variable = one_minus_advected_sq
other_name_weight_variable_component = 0
with the following parameters
hist_names=STRING, STRING, STRING, ...(comma separated names)The names of the histograms in this block. Will be used as prefix in the block as well as in the output file. All histograms will be written to the same output file with the “group” in the output corresponding to the histogram name.
NAME_ndim=INT(either1or2)Dimensionality of the histogram.
NAME_x_variable=STRING(variable name or special coordinate stringHIST_COORD_X1,HIST_COORD_X2,HIST_COORD_X3orHIST_COORD_R)Variable to be used as bin. If a variable name is given a component has to be specified, too, see next parameter. For a scalar variable, the component needs to be specified as
0anyway. If binning should be done by coordinate the special strings allow to bin by either one of the three dimensions or by radial distance from the origin.
NAME_x_variable_component=INTComponent index of the binning variable. Used/required only if a non-coordinate variable is used for binning.
NAME_x_edges_type=STRING(lin,log, orlist)How the bin edges are defined in the first dimension. For
linandlogdirect indexing is used to determine the bin, which is significantly faster than specifying the edges via alistas the latter requires a binary search.
NAME_x_edges_min=FLOATMinimum value (inclusive) of the bins in the first dim. Used/required only for
linandlogedge type.
NAME_x_edges_max=FLOATMaximum value (inclusive) of the bins in the first dim. Used/required only for
linandlogedge type.
NAME_x_edges_num_bins=INT(must be>=1)Number of equally spaced bins between min and max value in the first dim. Used/required only for
linandlogedge type.
NAME_x_edges_list=FLOAT,FLOAT,FLOAT,...(comma separated list of increasing values)Arbitrary definition of edge values with inclusive innermost and outermost edges. Used/required only for
listedge type.
NAME_y_edges...Same as the
NAME_x_edges...parameters except for being used in the second dimension forndim=2histograms.
NAME_accumulate=BOOL(trueorfalsedefault)Accumulate data that is outside the binning range in the outermost bins.
NAME_binned_variable=STRING(variable name orHIST_ONES)Variable to be binned. If a variable name is given a component has to be specified, too, see next parameter. For a scalar variable, the component needs to be specified as
0anyway. If sampling (i.e., counting the number of value inside a bin) is to be used the special stringHIST_ONEScan be set.
NAME_binned_variable_component=INTComponent index of the variable to be binned. Used/required only if a variable is binned and not
HIST_ONES.
NAME_weight_by_volume=BOOL(trueorfalse)Apply volume weighting to the binned variable. Can be used simultaneously with binning by a different variable. Note that this does not include any normalization (e.g., by total volume or the sum of the weight variable in question) and is left to the user during post processing.
NAME_weight_variable=STRINGVariable to be used as weight. Can be used together with volume weighting. For a scalar variable, the component needs to be specified as
0anyway.
NAME_weight_variable_component=INTComponent index of the variable to be used as weight.
Note, weighting by volume and variable simultaneously might seem counterintuitive, but easily allows for, e.g., mass-weighted profiles, by enabling weighting by volume and using a mass density field as additional weight variable.
In practice, a 1D histogram in the astrophysical context may look like (top panel from Fig 4 in Curtis et al 2023 ApJL 945 L13):
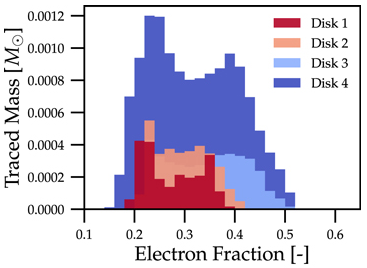
Translating this to the notation used for Parthenon histogram outputs means specifying for each histogram
the field containing the Electron fraction as
x_variable,the field containing the traced mass density as
binned_variable, andenable
weight_by_volume(to get the total traced mass).
Similarly, a 2D histogram (also referred to as phase plot) example may look like (from the yt Project documentation):
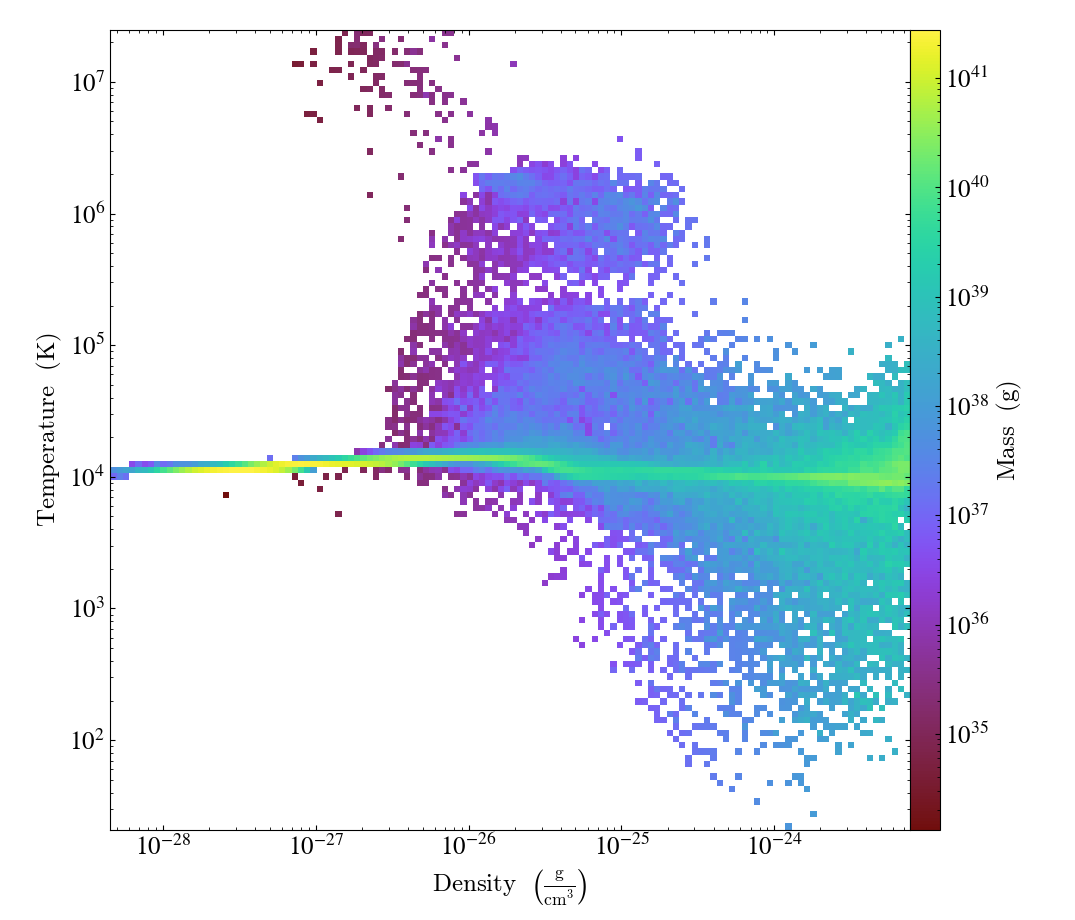
Translating this to the notation used for Parthenon histogram outputs means using
the field containing the density as
x_variable,the field containing the temperature as
y_variable,the field containing the mass density as
binned_variable, andenable
weight_by_volume(to get the total mass).
The following is a minimal example to plot a 1D and 2D histogram from the output file:
with h5py.File("parthenon.out8.histograms.00040.hdf", "r") as infile:
# 1D histogram
x = infile["myname/x_edges"][:]
y = infile["myname/data"][:]
plt.plot(x, y)
plt.show()
# 2D histogram
x = infile["other_name/x_edges"][:]
y = infile["other_name/y_edges"][:]
z = infile["other_name/data"][:].T # note the transpose here (so that the data matches the axis for the pcolormesh)
plt.pcolormesh(x,y,z,)
plt.show()
Ascent (optional)
Parthenon supports in situ visualization and analysis via the external
Ascent library.
Support for Ascent is disabled by default and must be enabled via PARTHENON_ENABLE_ASCENT=ON during configure.
In the input file, include a <parthenon/output*> block and specify file_type = ascent.
A dt parameter controls the frequency of outputs for simulations involving evolution.
Note that in principle Ascent can control its own output cadence (including
automated triggers).
If you want to call Ascent on every cycle, set dt to a value smaller than the actual simulation dt.
The mandatory actions_file parameter points to a separate file that defines
Ascent actions in .yaml or .json format, see
Ascent documentation for a complete list of options.
Parthenon currently only publishes cell-centered variables to Ascent.
Moreover, the published name of the field always starts with the base name (to avoid
name clashes between multiple fields that may have the same [component] labels).
If component label(s) are provided, they will be added as a suffix, e.g,.
basename_component-label for all variable types (even scalars).
Otherwise, an integer index is added for vectors/tensors with more than one component, i.e.,
vectors/tensors with a single component and without component labels will not contain a suffix.
The definition of component labels for variables is typically done by downstream codes
so that the downstream documentation should be consulted for more specific information.
A <parthenon/output*> block might look like:
<parthenon/output9>
file_type = ascent
dt = 1.0
actions_file = my_actions.yaml
see also the advection example input file and actions file.
Note by default “field filtering” is enabled for Ascent in Parthenon, i.e.,
only fields that are used in Ascent actions are published.
There may be cases, where Ascent cannot determine which fields it needs for
an action and will fail.
In this case, add an ascent_options.yaml file to the run directory containing:
field_filtering: false
to override at runtime. See Ascent documenation for more information.
Python scripts
The scripts/python folder includes scripts that may be useful for
visualizing or analyzing data in the .phdf files. The phdf.py
file defines a class to read in and query data. The movie2d.py
script shows an example of using this class, and also provides a
convenient means of making movies of 2D simulations. The script can be
invoked as
python3 /path/to/movie2d.py name_of_variable *.phdf
which will produce a png image per dump suitable for encoding into a
movie.
Visualization software
Both ParaView and
VisIt are
capable of opening and visualizing Parthenon graphics dumps. In both
cases, the .xdmf files should be opened. In ParaView, select the
“XDMF Reader” when prompted.
Warning
Currently parthenon face- and edge- centered data is not supported for ParaView and VisIt. However, our python tooling does support all mesh locations.
Preparing outputs for yt
Parthenon HDF5 outputs can be read with the python visualization library
yt as certain variables are named when
adding fields via StateDescriptor::AddField and
StateDescriptor::AddSparsePool. Variable names are added as a
std::vector<std::string> in the variable metadata. These labels are
optional and are only used for output to HDF5. 4D variables are named
with a list of names for each row while 3D variables are named with a
single name. For example, the following configurations are acceptable:
auto pkg = std::make_shared<StateDescriptor>("Hydro");
/* ... */
const int nhydro = 5;
std::vector<std::string> cons_labels(nhydro);
cons_labels[0]="Density";
cons_labels[1]="MomentumDensity1";
cons_labels[2]="MomentumDensity2";
cons_labels[3]="MomentumDensity3";
cons_labels[4]="TotalEnergyDensity";
Metadata m({Metadata::Cell, Metadata::Independent, Metadata::FillGhost},
std::vector<int>({nhydro}), cons_labels);
pkg->AddField("cons", m);
const int ndensity = 1;
std::vector<std::string> density_labels(ndensity);
density_labels[0]="Density";
m = Metadata({Metadata::Cell, Metadata::Derived}, std::vector<int>({ndensity}), density_labels);
pkg->AddField("dens", m);
const int nvelocity = 3;
std::vector<std::string> velocity_labels(nvelocity);
velocity_labels[0]="Velocity1";
velocity_labels[1]="Velocity2";
velocity_labels[2]="Velocity3";
m = Metadata({Metadata::Cell, Metadata::Derived}, std::vector<int>({nvelocity}), velocity_labels);
pkg->AddField("vel", m);
const int npressure = 1;
std::vector<std::string> pressure_labels(npressure);
pressure_labels[0]="Pressure";
m = Metadata({Metadata::Cell, Metadata::Derived}, std::vector<int>({npressure}), pressure_labels);
pkg->AddField("pres", m);
The yt frontend needs either the hydrodynamic conserved variables or
primitive compute derived quantities. The conserved variables must have
the names "Density", "MomentumDensity1", "MomentumDensity2",
"MomentumDensity3", "TotalEnergyDensity" while the primitive
variables must have the names "Density", "Velocity1",
"Velocity2", "Velocity3", "Pressure". Either of these sets
of variables must be named and present in the output, with the primitive
variables taking precedence over the conserved variables when computing
derived quantities such as specific thermal energy. In the above
example, including either "cons" or "dens", "vel", and
"pres" in the HDF5 output would allow yt to read the data.
Additional parameters can also be packaged into the HDF5 file to help
yt interpret the data, namely adiabatic index and code unit
information. These are identified by passing true as an optional
boolean argument when adding parameters via
StateDescriptor::AddParam. For example,
pkg->AddParam<double>("CodeLength", 100,true);
pkg->AddParam<double>("CodeMass", 1000,true);
pkg->AddParam<double>("CodeTime", 1,true);
pkg->AddParam<double>("AdibaticIndex", 5./3.,true);
pkg->AddParam<int>("IntParam", 0,true);
pkg->AddParam<std::string>("EquationOfState", "Adiabatic",true);
adds the parameters CodeLength, CodeMass, CodeTime,
AdiabaticIndex, IntParam, and EquationOfState to the HDF5
output. Currently, only int, float, and std::string
parameters can be included with the HDF5.
Code units can be defined for yt by including the parameters
CodeLength, CodeMass, and CodeTime, which specify the code
units used by Parthenon in terms of centimeters, grams, and seconds by
writing the parameters. In the above example, these parameters dictate
yt to interpret code lengths in the data in units of 100 centimeters
(or 1 meter per code unit), code masses in units of 1000 grams (or 1
kilogram per code units) and code times in units of seconds (or 1 second
per code time). Alternatively, this unit information can also be
supplied to the yt frontend when loading the data. If code units are
not defined in the HDF5 file or at load time, yt will assume that
the data is in CGS.
The adiabatic index can also be specified via the parameter
AdiabaticIndex, defined at load time for yt, or left as its
default 5./3..
For example, the following methods are valid to load data with yt
filename = "parthenon.out0.00000.phdf"
#Read units and adiabatic index from the HDF5 file or use defaults
ds = yt.load(filename)
#Specify units and adiabatic index explicitly
units_override = {"length_unit" : (100, "cm"),
"time_unit" : (1, "s"),
"mass_unit" : (1000,"g")}
ds = yt.load(filename,units_override=units_override,gamma=5./3.)
Currently, the yt frontend for Parthenon is hosted on the
parthenon-frontend branch of this yt fork. In
the future, the Parthenon frontend will be included in the main yt
repo.